Hieu Van Dong1,†, Dai Quang Trinh2,†, Giang Huong Thi Tran1, Thanh Thi Vu1,2, Thinh Hung Ba Nguyen1, Amonpun Rattanasrisomporn3, Dao Anh Tran Bui1, Jatuporn Rattanasrisomporn4
1Faculty of Veterinary Medicine, Vietnam National University of Agriculture
2Central Veterinary Medicine Joint Stock Company No. 5
3Kasetsart University, Thailand
A new study by a team from the Faculty of Veterinary Medicine, Vietnam National University of Agriculture, in collaboration with Central Veterinary Medicine Joint Stock Company No. 5 and Kasetsart University (Thailand), has identified the emergence of recombinant Duck circovirus (DuCV) strains circulating in several northern provinces of Vietnam during 2023–2024. This was an indication that the virus was continuing to evolve and could pose serious challenges to the health of domestic duck populations and the country’s waterfowl industry.
Warning from practical duck farming
Duck circovirus (DuCV) is a virus associated with stunting syndrome and feathering disorders in ducks, first discovered in Germany in 2003. This virus weakens the immune system, making ducks more susceptible to secondary infections such as parvovirus, E. coli, or Riemerella anatipestifer. In Vietnam, DuCV had been reported in both the north and the south since 2021, causing notable impacts on productivity and health.
Latest study and key findings
In this study, the authors collected 102 pooled visceral samples from ducks showing clinical symptoms in seven provinces/cities, including Thai Nguyen, Bac Giang, Hai Duong, Nghe An, Phu Tho, Hanoi, and Ha Nam. The results showed that 16 samples (35.56%) was positive for DuCV by PCR. Of these, nine representative samples were subjected to whole-genome sequencing using the Sanger method.
Notable findings
DuCV genomes ranged from 1992–1995 bp, with nucleotide similarity rates of 96.88–99.84%.
Phylogenetic analysis showed that all strains belonged to Genotype I, including subgroups Ia, Ib, and Ic, and were genetically closely related to DuCV strains recorded in China between 2019 and 2023.
Notably, genetic recombination events were identified in the Vietnam/VNUA-102/2023 and Vietnam/VNUA-225/2023 strains, confirmed by RDP5 and Simplot software.
The analysis of the Rep protein gene indicated negative selection pressure, suggesting the virus may be adapting more efficiently to its host.
Significance and recommendations
The study provided solid evidence of the sustained circulation and ongoing evolution of DuCV in Vietnam, in the context of no specific control measures available. The detection of recombinant strains raised concerns about the potential increase in virulence and transmissibility of the virus in the future.
This served as an urgent reminder for veterinary authorities and the duck farming community to strengthen epidemiological surveillance, ensure early diagnosis, and develop effective control programs for DuCV as well as its associated secondary infections.
Link to access the full text of the article: https://pmc.ncbi.nlm.nih.gov/articles/PMC12115742/
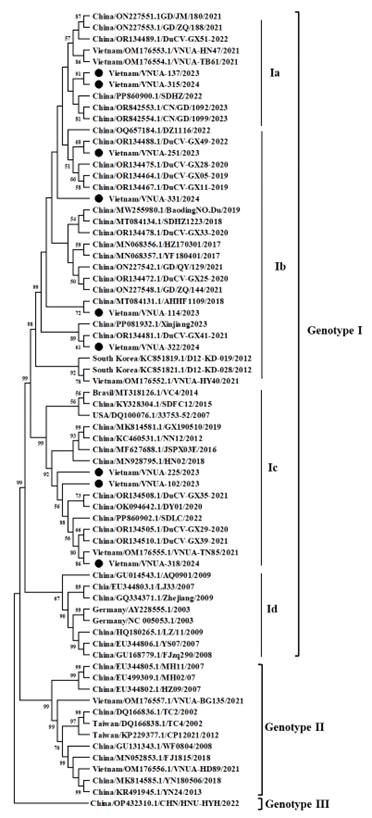
Figure 1. The phylogenetic tree of DuCV based on the complete genome sequences
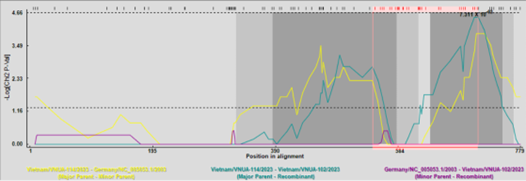
Figure 2. Detection of recombination events of full-length Cap gene sequences by the RDP 5 program
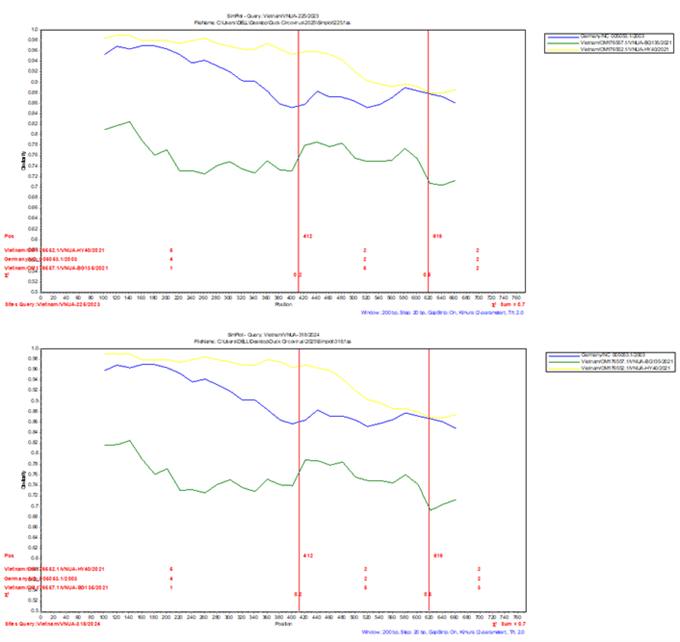
Figure 3. Detection of genetic recombination events using Simplot program